SNPs de novo predicted from
Atlantic Ocean and Mediterranean sea
Clic for zooming
Data
Description
All
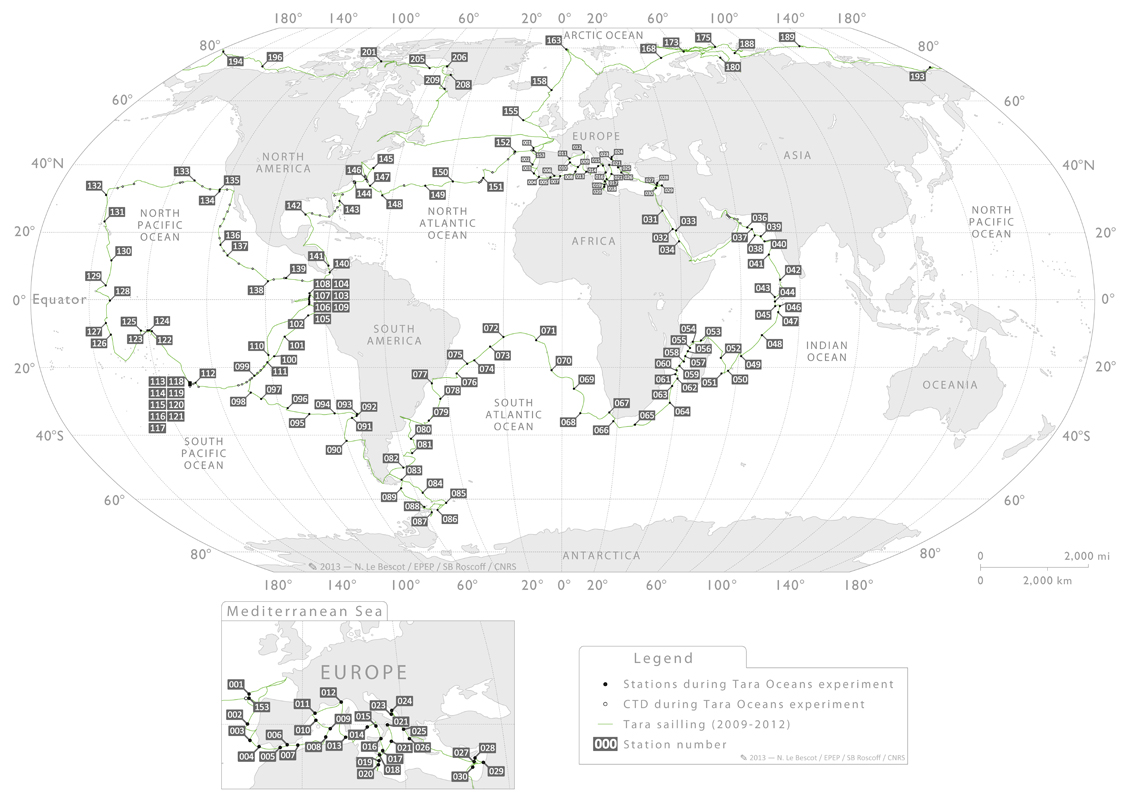
experiments provided here come from 53 distinct Tara Oceans stations. They correspond to stations numbers 4 to 30,
station numbers 66, 67, 68, 70, 72,76, 78, 80, 81, 82, 83, 84, 85, 86, 89, and
142 to 152. For each filter size variants were computed merging data from all
the 53 stations.
For each
filter size, following information and files are provided:
á Read file correspondence: .fa and .vcf files
refer to read sets as indices 1 to n (n=53). For each allele of each variant
and for each read set, these indices are used for designing the coverage (C1, C2, , Cn), and the phred quality (Q1, Q2, , Qn).
á A fasta file.
Each variant is provided as a couple of sequences. For each variant, the
following pieces of information are provided (among others)
o
Two
genomic sequences, distinct by SNP variants
o
The
rank: Variants with rank close to 0 are related to variants that do not
discriminate well read sets, while rank close to one design discriminative
variant (see DiscoSnp publication)
o
The
coverage per allele and per read set. The term coverage designs here the number of read mapped.
o
The
average phred quality of mapped reads.
á A vcf file. This file does not provide additional
information with respect to the fasta file. It is a VCF formatting of fasta
results. As these results were computed reference-free,
the mapping position of each variant simply designs the position where it
occurs in the sequence provided in the fasta file.
How to use data
The .vcf files provides a global view of all predicted variants.
It may serve to provide statistics per filter size or, more specifically,
statistics per station, by analyzing read coverages.
The .fa files can be used for downstream analyzes, in relation
with species of interest for which reference genome(s) is/are available. To
this aim, DiscoSnp propose
a script (runVCF_creator.sh) that maps variants contained in such .fa files to reference genomes, and provide a vcf file summing up mapped variants location pieces of
information.
Citation
If you use these data, please cite
Discovering Millions of Plankton Genomic Markers from the Atlantic
Ocean and the Mediterranean
Sea
Majda Arif, JŽrŽmy Gauthier, Kevin
Sugier, Daniele Iudicone, Olivier Jaillon,
Patrick Wincker, Pierre Peterlongo, Mohammed-Amin Madoui
Molecular Ecology
Resources, Wiley/Blackwell,
2018, pp.1-24. 〈10.1111/1755-0998.12985〉
Contact
pierre.peterlongo@inria.fr
Results per filter size
0.8 μm
to 5 μm
á Command: run_discoSnp++.sh -b 1 -r GGMM.fof
-p GGMM_all_set2 -k 51 -D 0 -t -P 3 -c 3
á fof file: fof_GGMM.zip
á Prediction statistics:
o
Number
of analysed reads: 11,434,589,112 reads
o
Number
of found variants: 6,920,311
o
SNPs:
¤
Transitions: 4,422,152 (63.9%)
¤
Transversions: 2,498,138 (36.1%)
á discoSnp:
o
Wall
clock computation time: 64h
o
Maximal
RAM memory: 107 GB
á Results:
o
Read
file correspondence: GGMM_all_set2_read_files_correspondence.txt
o
Fasta
file: GGMM_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.fa.gz
(1.0 GB)
o
VCF
file: GGMM_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.vcf.gz
(442 MB)
5 μm
to 20 μm
á Command: run_discoSnp++.sh -b 1 -r MMQQ.fof
–p MMQQ_all_set2 -k 51 -D 0 -t -P 3 -c 3
á fof file: fof_MMQQ.zip
á Prediction statistics:
o
Number
of analysed reads: 11,866,271,573 reads
o
Number
of found variants: 2,618,663
o
SNPs
¤
Transitions: 1,850,925
(58.3%)
¤
Transversions: 1,322,876 (41.7%)
á discoSnp:
o
Wall
clock computation time: 60 h
o
Maximal
RAM memory: 107 GB
á Results:
o
Read
file correspondence: MMQQ_all_set2_read_files_correspondence.txt
o
Fasta
file: MMQQ_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.fa.gz
(360 MB)
o
VCF
file: MMQQ_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.vcf.gz
(157 MB)
20 μm
to 180 μm
á Command: run_discoSnp++.sh -b 1 -r QQSS.fof
–p QQSS_all_set2 -k 51 -D 0 -t -P 3 -c 3
á fof file: fof_QQSS.zip
á Prediction statistics:
o
Number
of analysed reads: reads: 11,223,993,138 reads
o
Number
of found variants: 5,208,905
o
SNPs
¤
Transitions: 3,763,631
(60.6%)
¤
Transversions: 2,443,478 (39.4)
á discoSnp:
o
Wall
clock computation time: 105 h
o
Maximal
RAM memory: 110 GB
á Results:
o
Read
file correspondence: QQSS_all_set2_read_files_correspondence.txt
o
Fasta
file: QQSS_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.fa.gz
(1.1 GB)
o
VCF
file: QQSS_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.vcf.gz
(533 MB)
180 μm
to 2000 μm
á Command: run_discoSnp++.sh -b 1 -r SSUU.fof
–p SSUU_all_set2 -k 51 -D 0 -t -P 3 -c 3
á fof file: fof_SSUU.zip
á Prediction statistics:
o
Number
of analysed reads: reads: 11,169,165,660 reads
o
Number
of found variants: 6,230,835
o
SNPs
¤
Transitions: 4,382,514
(58.5%)
¤
Transversions: 3,114,523 (41.5%)
á discoSnp:
o
Wall
clock computation time: 124 h
o
Maximal
RAM memory: 120 GB
á Results:
o
Read
file correspondence: SSUU_all_set2_read_files_correspondence.txt
o
Fasta
file: SSUU_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.fa.gz
(1.1 GB)
o
VCF
file: SSUU_all_set2_k_51_c_3_D_0_P_3_b_1_coherent.vcf.gz
(499 MB)